Workshop on R
May 2018
Hosted by Virginia Education Science Training (VEST) Program at UVA
Overview Schedule Getting started Modules Data
High performance R with Rcpp
R is really powerful, but for some tasks can be very slow. As data get larger and scripts require more complex algorithms and demanding tasks, even small bits of sluggishness become huge in the aggregate. Compiled languages such as C and C++ can be much faster than R, but require greater programming skill. While learning a compiled language is useful for many researchers, the upfront time needed to learn is often too steep for any given project.
Fortunately, the Rcpp package bridges the gap between the two worlds. While there is a bit of a learning curve for Rcpp, which is based on C++, much R code can be, with few modifications, adjusted to become Rcpp code and run much faster for complex tasks.
This module will give one example of such a speed up. In my own research, I’ve computed the Great Circle (“as the crow flies”) distance between colleges and various geographic centroids many times using either the Haversine or Vincenty formula. But with over 7,500 colleges and universities in the United States, measuring the distance from each county centroid (around 3,000) became slow enough. Doing the same for all census tracks (around 70,000) and census block groups (over 200,000) was almost impossible. With small modifications to base R formula functions, however, I was able to convert them to compiled Rcpp function which run much, much faster.
## libraries
library(tidyverse)
library(Rcpp)
library(microbenchmark)
Load data
This module uses two data frames. The first has the names and locations of all colleges with a physical campus in 2015. The second has the locations of every census block group in the United States from the 2010 Census.
## load data
load('../data/hp_rcpp.Rdata')
College locations
Here’s a quick peek at the college location data (around 7,600 institutions).
## college locations
col_df
# A tibble: 7,647 x 5
unitid instnm fips5 lon lat
<int> <chr> <chr> <dbl> <dbl>
1 100654 Alabama A & M University 01089 -86.6 34.8
2 100663 University of Alabama at Birmingham 01073 -86.8 33.5
3 100690 Amridge University 01101 -86.2 32.4
4 100706 University of Alabama in Huntsville 01089 -86.6 34.7
5 100724 Alabama State University 01101 -86.3 32.4
6 100733 University of Alabama System Office 01125 -87.5 33.2
7 100751 The University of Alabama 01125 -87.5 33.2
8 100760 Central Alabama Community College 01123 -85.9 32.9
9 100812 Athens State University 01083 -87.0 34.8
10 100830 Auburn University at Montgomery 01101 -86.2 32.4
# ... with 7,637 more rows
Census block group locations
And here’s the census block group location data (around 217,000 block groups).
## census block group locations
cbg_df
# A tibble: 217,740 x 4
fips11 pop lon lat
<chr> <int> <dbl> <dbl>
1 010010201001 698 -86.5 32.5
2 010010201002 1214 -86.5 32.5
3 010010202001 1003 -86.5 32.5
4 010010202002 1167 -86.5 32.5
5 010010203001 2549 -86.5 32.5
6 010010203002 824 -86.5 32.5
7 010010204001 944 -86.4 32.5
8 010010204002 1937 -86.4 32.5
9 010010204003 935 -86.4 32.5
10 010010204004 570 -86.4 32.5
# ... with 217,730 more rows
Compute great circle distance with Haversine
The Haversine
formula
is a fairly straightforward trigonometric problem. Since the coordinates
in the data are in latitude and longitude and the formula requires
radians, a quick helper function deg_to_rad() is used to make the
conversion.
## convert degrees to radians
deg_to_rad <- function(degree) {
m_pi <- 3.141592653589793238462643383280
return(degree * m_pi / 180)
}
## compute Haversine distance between two points
dist_haversine <- function(xlon, xlat, ylon, ylat) {
## radius of Earth in meters
e_r <- 6378137
## return 0 if same point
if (xlon == ylon & xlat == xlon) { return(0) }
## convert degrees to radians
xlon = deg_to_rad(xlon)
xlat = deg_to_rad(xlat)
ylon = deg_to_rad(ylon)
ylat = deg_to_rad(ylat)
## haversine distance formula
d1 <- sin((ylat - xlat) / 2)
d2 <- sin((ylon - xlon) / 2)
return(2 * e_r * asin(sqrt(d1^2 + cos(xlat) * cos(ylat) * d2^2)))
}
With the formula, we can compute the distance in meters between the first census block group and the first college in the data set pretty quickly.
## store first census block group point (x) and first college point (y)
xlon <- cbg_df[[1, 'lon']]
xlat <- cbg_df[[1, 'lat']]
ylon <- col_df[[1, 'lon']]
ylat <- col_df[[1, 'lat']]
## test single distance function
d <- dist_haversine(xlon, xlat, ylon, ylat)
d
[1] 258212.3
Quick exercise
Find the coordinates of two places you know the distance between pretty well (say, your hometown and where you live now or your first college or the nearest big city). Compute the distance and compare to Google’s driving distance. It should be shorter (crows fly very straight), but similar. You may want to convert the meters to kilometers or miles. As always Google is your friend for all these steps.
Many to many distance matrix
Now that we have a core function, let’s write a larger function that can take many input points, many output points, and compute the distances between them.
## compute many to many distances and return matrix
dist_mtom <- function(xlon, # vector of starting longitudes
xlat, # vector of starting latitudes
ylon, # vector of ending longitudes
ylat, # vector of ending latitudes
x_names, # vector of starting point names
y_names) { # vector of ending point names
## init output matrix (n X k)
n <- length(xlon)
k <- length(ylon)
mat <- matrix(NA, n, k)
## double loop through each set of points to get all combinations
for(i in 1:n) {
for(j in 1:k) {
## compute distance using core function
mat[i,j] <- dist_haversine(xlon[i], xlat[i], ylon[j], ylat[j])
}
}
## add row and column names
rownames(mat) <- x_names
colnames(mat) <- y_names
return(mat)
}
Let’s test it with a subset of ten starting points.
## test matrix (limit to only 10 starting points)
distmat <- dist_mtom(cbg_df$lon[1:10], cbg_df$lat[1:10],
col_df$lon, col_df$lat,
cbg_df$fips11[1:10], col_df$unitid)
## show
distmat[1:5,1:5]
100654 100663 100690 100706 100724
010010201001 258212.3 119349.8 31495.73 251754.3 21138.09
010010201002 256255.2 117447.3 32284.61 249798.4 22263.60
010010202001 256775.4 118210.3 31047.25 250348.1 21059.86
010010202002 258084.9 119554.9 30210.67 251665.2 20017.60
010010203001 256958.5 118703.3 29758.10 250567.0 19905.92
Quick exercise
Can you find the minimum distance for each starting point? What’s the name of the nearest end point?
Nearest end point
Rather than return a full matrix of distances that we then need to
evaluate to find the shortest distance, let’s write a new function,
dist_min(), that will take two vectors of points again, but only
return the closest point in the second group, with distance, for each
point in the first group.
## compute and return minimum distance along with name
dist_min <- function(xlon, # vector of starting longitudes
xlat, # vector of starting latitudes
ylon, # vector of ending longitudes
ylat, # vector of ending latitudes
x_names, # vector of starting point names
y_names) { # vector of ending point names
## NB: lengths: x coords == x names && y coords == y_names
n <- length(xlon)
k <- length(ylon)
minvec_name <- vector('character', n)
minvec_meter <- vector('numeric', n)
## init temporary vector for distances between one x and all ys
tmp <- vector('numeric', k)
## give tmp names of y vector
names(tmp) <- y_names
## loop through each set of starting points
for(i in 1:n) {
for(j in 1:k) {
tmp[j] <- dist_haversine(xlon[i], xlat[i], ylon[j], ylat[j])
}
## add to output matrix
minvec_name[i] <- names(which.min(tmp))
minvec_meter[i] <- min(tmp)
}
return(data.frame('fips11' = x_names,
'unitid' = minvec_name,
'meters' = minvec_meter,
stringsAsFactors = FALSE))
}
Let’s test it out.
## test matrix (limit to only 10 starting points)
mindf <- dist_min(cbg_df$lon[1:10], cbg_df$lat[1:10],
col_df$lon, col_df$lat,
cbg_df$fips11[1:10], col_df$unitid)
## show
mindf
fips11 unitid meters
1 010010201001 101471 15785.88
2 010010201002 101471 14228.69
3 010010202001 101471 13949.21
4 010010202002 101471 14870.35
5 010010203001 101471 13362.81
6 010010203002 101471 14389.79
7 010010204001 101471 12704.73
8 010010204002 101471 13270.80
9 010010204003 101471 14102.44
10 010010204004 101471 14756.51
Quick exercise
How long will it take to find the closest college to each census block group? Use
system.time()and extrapolate to make a best guess.
Rcpp
These worked well, but we only did 10 starting points. We need to find it for over 200,000 starting points. What we need is to convert the base R functions into Rcpp functions.
Below, the full script, dist_func.cpp, is discussed in pieces.
Front matter
The head of an Rcpp script is like an R script in that this is place to
#include other header files. This is akin to having library(...) in
an R script. At the very least, an Rcpp script needs to include it’s own
header files, Rcpp.h.
Next, we define preprocessor replacements. In the process of compiling
the script, that is, turning the Rcpp code that’s human readable into
machine-readable byte code, the preprocessor first runs through the
script. One job is to replace any #define directive with the value it
has been given. In our code, that means that anywhere in the script that
e_r is found, 6378137.0 literally replaces it.
Finally, we load the Rcpp namespace with using namespace Rcpp;. This
means that we don’t have to keep repeating Rcpp::<...> before every
function we want. Sometimes, it’s better to do it the long way,
especially if you are using functions from multiple libraries that may
overlap, but we’re okay doing it this way this time.
// header files to include
#include <Rcpp.h>
// preprocessor replacements
#define e_r 6378137.0
#define m_pi 3.141592653589793238462643383280
// use Rcpp namespace to avoid Rcpp::<...> repetition
using namespace Rcpp;
Utility functions
First, we have the two utility functions to convert degrees to radians and to compute the Haversine distance between two points. A few things to note right away:
- Rcpp comments use
//or/* ... */ - Lines must end in a semi-colon,
; - Variables are strongly typed
When a language is strongly typed, the user must tell the compiler the
variable type, like double, int, etc. R will guess what you want and
change on fly. This is a nice interactive feature, but is part of what
makes R slow sometimes. By strongly typing a variable, the computer
knows right away what you want and doesn’t waste time or memory making
adjustments.
A few other things to notice:
doublesshould end in a.or.0, otherwise they are assumed to be integers; dividing by integers can have weird outcomes so unless you are REALLY sure you want an integer, a floating point number like adoubleis probably what you want (seedegree * m_pi / 180.0)- The variable type before the function name tells the compiler what form the function output will take
- Function arguments must also be typed
Finally, for the function to be available to you as a user, you must put
// [[Rcpp::export]] just before it.
// convert degrees to radians
// [[Rcpp::export]]
double deg_to_rad_rcpp(double degree) {
return(degree * m_pi / 180.0);
}
// compute Haversine distance between two points
// [[Rcpp::export]]
double dist_haversine_rcpp(double xlon,
double xlat,
double ylon,
double ylat) {
// return 0 if same point
if (xlon == ylon && xlat == xlon) return 0.0;
// convert degrees to radians
xlon = deg_to_rad_rcpp(xlon);
xlat = deg_to_rad_rcpp(xlat);
ylon = deg_to_rad_rcpp(ylon);
ylat = deg_to_rad_rcpp(ylat);
// haversine distance formula
double d1 = sin((ylat - xlat) / 2.0);
double d2 = sin((ylon - xlon) / 2.0);
return 2.0 * e_r * asin(sqrt(d1*d1 + cos(xlat) * cos(ylat) * d2*d2));
}
Many to many distance matrix
Moving to the main functions, notice that base R function has been changed only slightly. For the most part, the difference is that variables, argument, and function return must be typed.
A few other differences to note:
- the length of a vector is measured with
.size() - loops in C++ take the form (
; ; ), which works as "start `i` as 0, check if `i` is less than the number of starting points, `n`, add 1 to `i`, then run the loop; if the check fails, skip the loop.
// compute many to many distances and return matrix
// [[Rcpp::export]]
NumericMatrix dist_mtom_rcpp(NumericVector xlon,
NumericVector xlat,
NumericVector ylon,
NumericVector ylat,
CharacterVector x_names,
CharacterVector y_names) {
// init output matrix (x X y)
int n = xlon.size();
int k = ylon.size();
NumericMatrix distmat(n,k);
// double loop through each set of points to get all combinations
for(int i = 0; i < n; i++) {
for(int j = 0; j < k; j++) {
distmat(i,j) = dist_haversine_rcpp(xlon[i],xlat[i],ylon[j],ylat[j]);
}
}
// add row and column names
rownames(distmat) = x_names;
colnames(distmat) = y_names;
return distmat;
}
Nearest end point
Again the Rcpp version of this script is almost identical to the base R
version, just with variable typing added. Since we’re returning a data
frame (Rcpp version: DataFrame), we have to create it with
DataFrame::create() as we return it.
// compute and return minimum distance along with name
// [[Rcpp::export]]
DataFrame dist_min_rcpp(NumericVector xlon,
NumericVector xlat,
NumericVector ylon,
NumericVector ylat,
CharacterVector x_names,
CharacterVector y_names) {
// init output matrix (x X 3)
int n = xlon.size();
int k = ylon.size();
CharacterVector minvec_name(n);
NumericVector minvec_meter(n);
NumericVector tmp(k);
// loop through each set of starting points
for(int i = 0; i < n; i++) {
for(int j = 0; j < k; j++) {
tmp[j] = dist_haversine_rcpp(xlon[i],xlat[i],ylon[j],ylat[j]);
}
// add to output matrix
minvec_name[i] = y_names[which_min(tmp)];
minvec_meter[i] = min(tmp);
}
// return created data frame
return DataFrame::create(Named("fips11") = x_names,
Named("unitid") = minvec_name,
Named("meters") = minvec_meter,
_["stringsAsFactors"] = false);
}
Source the Rcpp file
To compile the Rcpp scripts and have the functions available to us in
our R script or session, we need to read in the script with
sourceCpp(). This works much like regular source() works, except for
Rcpp files. We’ll add the argument rebuild = TRUE so that if need to
go back and adjust the source code during a session, it will be rebuilt
when we call sourceCpp() again.
Compiling code can take a while. In essence, we’re trading some time now for faster speed down the road. This is why compiled code isn’t great for interactive coding sessions or for small tasks. But our code isn’t complex and compiles rather quickly.
## source Rcpp code
sourceCpp('../scripts/dist_func.cpp', rebuild = TRUE)
Quick comparisons
Now that we have both versions of our distance measuring functions, we should compare the time it takes both to run. But first, let’s make sure that they give the same results.
Single distance
d_Rcpp <- dist_haversine_rcpp(xlon, xlat, ylon, ylat)
## show
d_Rcpp
[1] 258212.3
## compare
identical(d, d_Rcpp)
[1] TRUE
For one point, our dist_haversine() and dist_haversine_rcpp() give
the same result.
Many to many
Next, we’ll compare our many to many matrix, making sure we get the same values.
distmat_Rcpp <- dist_mtom_rcpp(cbg_df$lon[1:10],
cbg_df$lat[1:10],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:10],
col_df$unitid)
## show
distmat_Rcpp[1:5,1:5]
100654 100663 100690 100706 100724
010010201001 258212.3 119349.8 31495.73 251754.3 21138.09
010010201002 256255.2 117447.3 32284.61 249798.4 22263.60
010010202001 256775.4 118210.3 31047.25 250348.1 21059.86
010010202002 258084.9 119554.9 30210.67 251665.2 20017.60
010010203001 256958.5 118703.3 29758.10 250567.0 19905.92
## compare
all.equal(distmat, distmat_Rcpp)
[1] TRUE
Again, we get the same values (within some very very small floating point rounding error).
Minimum distances
Finally, we’ll compare the minimum distance function.
mindf_Rcpp <- dist_min_rcpp(cbg_df$lon[1:10],
cbg_df$lat[1:10],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:10],
col_df$unitid)
## show
mindf_Rcpp
fips11 unitid meters
1 010010201001 101471 15785.88
2 010010201002 101471 14228.69
3 010010202001 101471 13949.21
4 010010202002 101471 14870.35
5 010010203001 101471 13362.81
6 010010203002 101471 14389.79
7 010010204001 101471 12704.73
8 010010204002 101471 13270.80
9 010010204003 101471 14102.44
10 010010204004 101471 14756.51
## compare
all.equal(mindf, mindf_Rcpp)
[1] TRUE
And once more, the same results. We’re now ready to compare speeds!
Benchmarks
To compare really small differences in time, we’ll use the
microbenchmark
package. Aside from being accurate at small time scales, it makes
comparisons based on multiple runs and can plot the differences using
the autoplot() function.
First, we’ll compare the core dist_haversine*() functions.
## use microbenchmark to compare
tm_single <- microbenchmark(base_R = dist_haversine(xlon, xlat, ylon, ylat),
Rcpp = dist_haversine_rcpp(xlon, xlat, ylon, ylat),
times = 1000L)
## results
tm_single
Unit: microseconds
expr min lq mean median uq max neval
base_R 2.746 2.845 2.935603 2.903 2.9700 8.242 1000
Rcpp 1.932 2.044 3.222372 2.134 2.2445 1066.782 1000
## plot
autoplot(tm_single)
Coordinate system already present. Adding new coordinate system, which will replace the existing one.
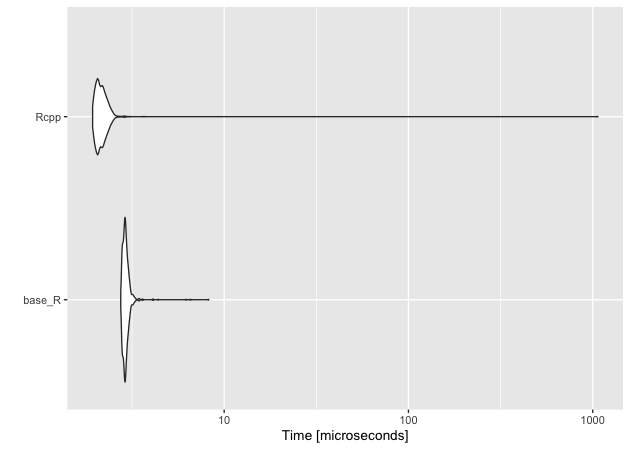
Comparing dist_haversine() with dist_haversine_rcpp(), the compiled
version isn’t that much faster. Considering we’re on the scale of
microseconds, it really isn’t that much faster.
## time for base R to do many to many with 100 starting points
system.time(dist_mtom(cbg_df$lon[1:100],
cbg_df$lat[1:100],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:100],
col_df$unitid))
user system elapsed
2.525 0.012 2.539
## ...and now Rcpp version
system.time(dist_mtom_rcpp(cbg_df$lon[1:100],
cbg_df$lat[1:100],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:100],
col_df$unitid))
user system elapsed
0.045 0.000 0.046
## compare just 10 many to many
tm_mtom <- microbenchmark(base_R = dist_mtom(cbg_df$lon[1:10],
cbg_df$lat[1:10],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:10],
col_df$unitid),
Rcpp = dist_mtom_rcpp(cbg_df$lon[1:10],
cbg_df$lat[1:10],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:10],
col_df$unitid),
times = 100L)
## results
tm_mtom
Unit: milliseconds
expr min lq mean median uq max
base_R 221.641507 227.737364 236.004614 233.302698 238.464247 308.327211
Rcpp 3.101263 3.511224 3.764691 3.746062 4.003859 4.796772
neval
100
100
## plot
autoplot(tm_mtom)
Coordinate system already present. Adding new coordinate system, which will replace the existing one.
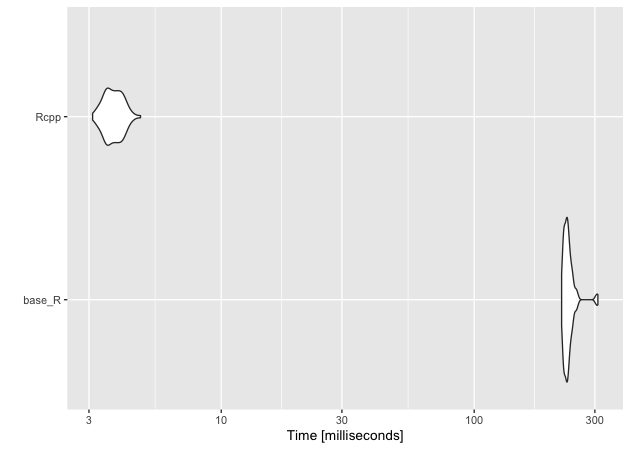
Where we start to see speed improvements in the main function. The compiled version of the many to many function is nearly two orders of magnitude faster!
## time for base R to do many to many with 100 starting points
system.time(dist_min(cbg_df$lon[1:100],
cbg_df$lat[1:100],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:100],
col_df$unitid))
user system elapsed
2.478 0.005 2.486
## ...and now Rcpp version
system.time(dist_min_rcpp(cbg_df$lon[1:100],
cbg_df$lat[1:100],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:100],
col_df$unitid))
user system elapsed
0.047 0.001 0.048
## compare just 10 min
tm_min <- microbenchmark(base_R = dist_min(cbg_df$lon[1:10],
cbg_df$lat[1:10],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:10],
col_df$unitid),
Rcpp = dist_min_rcpp(cbg_df$lon[1:10],
cbg_df$lat[1:10],
col_df$lon,
col_df$lat,
cbg_df$fips11[1:10],
col_df$unitid),
times = 100)
## results
tm_min
Unit: milliseconds
expr min lq mean median uq max
base_R 215.037837 239.062711 247.171698 246.488845 252.828805 317.801239
Rcpp 3.382741 4.003108 4.223458 4.187064 4.423666 5.330061
neval
100
100
## plot
autoplot(tm_min)
Coordinate system already present. Adding new coordinate system, which will replace the existing one.
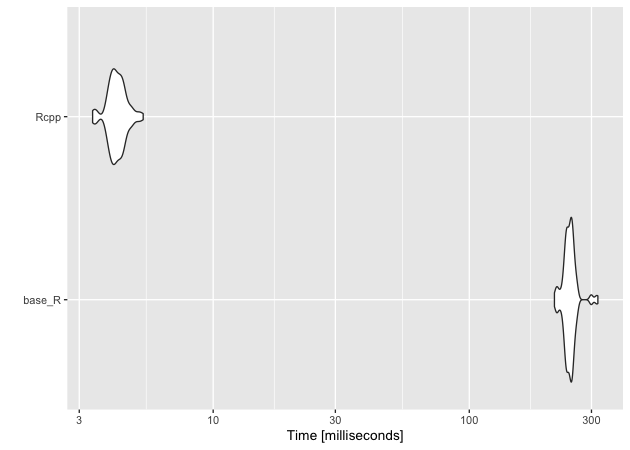
Similarly, the compiled minimum distance function is much faster than the base R function.
In this case as well as the former, we should note that we aren’t comparing fully optimized versions of either function. That said, while it’s possible that the base R functions could be sped up, neither is likely to come close to matching the speed of the compiled Rcpp versions.
Full run for Rcpp version
Throughout, we’ve been running the functions on a reduced starting data set. But how long does it take to find the nearest college to each census block? Let’s find out!
## find minimum
system.time(full_min <- dist_min_rcpp(cbg_df$lon,
cbg_df$lat,
col_df$lon,
col_df$lat,
cbg_df$fips11,
col_df$unitid))
user system elapsed
75.987 0.144 76.301
## show
full_min %>% tbl_df()
# A tibble: 217,740 x 3
fips11 unitid meters
<chr> <chr> <dbl>
1 010010201001 101471 15786.
2 010010201002 101471 14229.
3 010010202001 101471 13949.
4 010010202002 101471 14870.
5 010010203001 101471 13363.
6 010010203002 101471 14390.
7 010010204001 101471 12705.
8 010010204002 101471 13271.
9 010010204003 101471 14102.
10 010010204004 101471 14757.
# ... with 217,730 more rows
Just a little over two minutes to compute 217,000 by 7,700 distances, finding and storing the minimally distance college along with its distance!
Not-so quick exercise
Below is a function that computes the great circle distance using Vincenty’s formula. It’s more accurate than the haversine version, but can be much more computationally intensive. Try to convert the base R function into an Rcpp function. You’ll need to start a new script and then use
sourceCpp()to read it in and test. Once you’ve got, substitute the respective Vincenty formula functions into thedist_min_*()functions and compare times.A few things to keep in mind:
1. You’ll need to declare your variables and types (lot’s ofdouble); don’t forget that double numbers need a decimal, otherwise C++ thinks they are integers.
2. Don’t forget your semi-colon line endings!
3.abs()in C++ isfabs()
4. Remember:a^2 = a * a
## base R distance function using Vincenty formula
dist_vincenty <- function(xlon,
xlat,
ylon,
ylat) {
## return 0 if same point
if (xlon == ylon && xlat == ylat) { return(0) }
## convert degrees to radians
xlon <- deg_to_rad(xlon)
xlat <- deg_to_rad(xlat)
ylon <- deg_to_rad(ylon)
ylat <- deg_to_rad(ylat)
## ---------------------------------------------------
## https:##en.wikipedia.org/wiki/Vincenty%27s_formulae
## ---------------------------------------------------
## some constants
a <- 6378137
f <- 1 / 298.257223563
b <- (1 - f) * a
U1 <- atan((1 - f) * tan(xlat))
U2 <- atan((1 - f) * tan(ylat))
sinU1 <- sin(U1)
sinU2 <- sin(U2)
cosU1 <- cos(U1)
cosU2 <- cos(U2)
L <- ylon - xlon
lambda <- L # initial value
## set up loop
iters <- 100 # no more than 100 loops
tol <- 1.0e-12 # tolerance level
again <- TRUE
while (again) {
## sin sigma
sinLambda <- sin(lambda)
cosLambda <- cos(lambda)
p1 <- cosU2 * sinLambda
p2 <- cosU1 * sinU2 - sinU1 * cosU2 * cosLambda
sinsig <- sqrt(p1^2 + p2^2)
## cos sigma
cossig <- sinU1 * sinU2 + cosU1 * cosU2 * cosLambda
## plain ol' sigma
sigma <- atan2(sinsig, cossig)
## sin alpha
sina <- cosU1 * cosU2 * sinLambda / sinsig
## cos^2 alpha
cos2a <- 1 - (sina * sina)
## cos 2*sig_m
cos2sigm <- cossig - 2 * sinU1 * sinU2 / cos2a
## C
C <- f / 16 * cos2a * (4 + f * (4 - 3 * cos2a))
## store old lambda
lambdaOld <- lambda
## update lambda
lambda <- L + (1 - C) * f * sina *
(sigma + C * sinsig * (cos2sigm + C * cossig *
(-1 + 2 * cos2sigm^2)))
## subtract from iteration total
iters <- iters - 1
## go again if lambda diff is > tolerance and still have iterations
again <- (abs(lambda - lambdaOld) > tol && iters > 0)
}
## if iteration count runs out, stop and send message
if (iters == 0) {
stop("Failed to converge!", call. = FALSE)
}
else {
## u^2
Usq <- cos2a * (a^2 - b^2) / (b^2)
## A
A <- 1 + Usq / 16384 * (4096 + Usq * (-768 + Usq * (320 - 175 * Usq)))
## B
B <- Usq / 1024 * (256 + Usq * (-128 + Usq * (74 - 47 * Usq)))
## delta sigma
dsigma <- B * sinsig *
(cos2sigm + B / 4 *
(cossig * (-1 + 2 * cos2sigm^2)
- B / 6 * cos2sigm * (-3 + 4 * sinsig^2)
* (-3 + 4 * cos2sigm^2)))
## return the distance
return(b * A * (sigma - dsigma))
}
}
